Your new post is loading...
ABSTRACT
In bacteria, the chromosome is folded into a structure called the nucleoid. This compaction must facilitate and maintain key biological processes such as gene expression and DNA transactions (replication, recombination, repair, segregation). Chromosome and chromatin 3D-organization in bacteria has been a puzzle for decades. Chromosome conformation capture coupled to deep sequencing (HiC) in combination with other ‘omics’ approaches has allowed dissection of the structural layers that shape bacterial chromosome organization, from DNA topology to global chromosome architecture. Here we review the latest findings in the field and discuss the main features of bacterial genome folding.
Abstract
Members of the alphaproteobacterial order Rhodobacterales are metabolically diverse and highly abundant in the ocean. They are becoming increasingly interesting for marine biotechnology, due to their ecological adaptability, wealth of versatile low-copy-number plasmids, and their ability to produce secondary metabolites. However, molecular tools for engineering strains of this bacterial lineage are limited. Here, we expand the genetic toolbox by establishing standardized, modular repABC-based plasmid vectors of four well-characterized compatibility groups from the Roseobacter group applicable in the Rhodobacterales, and likely in further alphaproteobacterial orders (Hyphomicrobiales, Rhodospirillales, Caulobacterales). We confirmed replication of these newly constructed pABC vectors in two members of Rhodobacterales, namely, Dinoroseobacter shibae DFL 12 and Rhodobacter capsulatus B10S, as well as in two members of the alphaproteobacterial order Hyphomicrobiales (synonym: Rhizobiales; Ensifer meliloti 2011 and “Agrobacterium fabrum” C58). Maintenance of the pABC vectors in the biotechnologically valuable orders Rhodobacterales and Hyphomicrobiales facilitates the shuttling of genetic constructs between alphaproteobacterial genera and orders. Additionally, plasmid replication was verified in one member of Rhodospirillales (Rhodospirillum rubrum S1) as well as in one member of Caulobacterales (Caulobacter vibrioides CB15N). The modular construction of pABC vectors and the usage of four compatible replication systems, which allows their coexistence in a host cell, are advantageous features for future implementations of newly designed synthetic pathways. The vector applicability was demonstrated by functional complementation of a nitrogenase mutant phenotype by two complementary pABC-based plasmids in R. capsulatus.
Abstract
There is increasing evidence that interactions between microbes and their hosts not only play a role in determining health and disease but also in emotions, thought, and behavior. Built environments greatly influence microbiome exposures because of their built-in highly specific microbiomes coproduced with myriad metaorganisms including humans, pets, plants, rodents, and insects. Seemingly static built structures host complex ecologies of microorganisms that are only starting to be mapped. These microbial ecologies of built environments are directly and interdependently affected by social, spatial, and technological norms. Advances in technology have made these organisms visible and forced the scientific community and architects to rethink gene–environment and microbe interactions respectively. Thus, built environment design must consider the microbiome, and research involving host–microbiome interaction must consider the built-environment. This paradigm shift becomes increasingly important as evidence grows that contemporary built environments are steadily reducing the microbial diversity essential for human health, well-being, and resilience while accelerating the symptoms of human chronic diseases including environmental allergies, and other more life-altering diseases. New models of design are required to balance maximizing exposure to microbial diversity while minimizing exposure to human-associated diseases. Sustained trans-disciplinary research across time (evolutionary, historical, and generational) and space (cultural and geographical) is needed to develop experimental design protocols that address multigenerational multispecies health and health equity in built environments.
ABSTRACT
Bacteria often experience nutrient limitation in nature and the laboratory. While exponential and stationary growth phases are well characterized in the model bacterium Escherichia coli, little is known about what transpires inside individual cells during the transition between these two phases. Through quantitative cell imaging, we found that the position of nucleoids and cell division sites becomes increasingly asymmetric during transition phase. These asymmetries were coupled with spatial reorganization of proteins, ribosomes, and RNAs to nucleoid-centric localizations. Results from live-cell imaging experiments, complemented with genetic and 13C whole-cell nuclear magnetic resonance spectroscopy studies, show that preferential accumulation of the storage polymer glycogen at the old cell pole leads to the observed rearrangements and asymmetric divisions. In vitro experiments suggest that these phenotypes are likely due to the propensity of glycogen to phase separate in crowded environments, as glycogen condensates exclude fluorescent proteins under physiological crowding conditions. Glycogen-associated differences in cell sizes between strains and future daughter cells suggest that glycogen phase separation allows cells to store large glucose reserves without counting them as cytoplasmic space.
Abstract
MucR belongs to a large protein family whose members regulate the expression of virulence and symbiosis genes in α-proteobacteria species. This protein and its homologs were initially studied as classical transcriptional regulators mostly involved in repression of target genes by binding their promoters. Very recent studies have led to the classification of MucR as a new type of Histone-like Nucleoid Structuring (H-NS) protein. Thus this review is an effort to put together a complete and unifying story demonstrating how genetic and biochemical findings on MucR suggested that this protein is not a classical transcriptional regulator, but functions as a novel type of H-NS-like protein, which binds AT-rich regions of genomic DNA and regulates gene expression.
Abstract
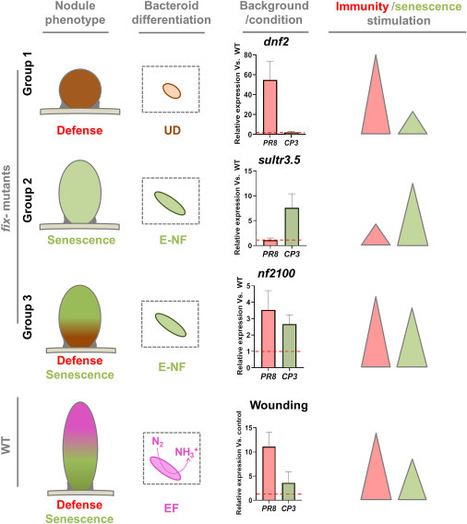
Immunity and senescence play a crucial role in the functioning of the legume symbiotic nodules. The miss-regulation of one of these processes compromises the symbiosis leading to death of the endosymbiont and the arrest of the nodule functioning. The relationship between immunity and senescence has been extensively studied in plant organs where a synergistic response can be observed. However, the interplay between immunity and senescence in the symbiotic organ is poorly discussed in the literature and these phenomena are often mixed up. Recent studies revealed that the cooperation between immunity and senescence is not always observed in the nodule, suggesting complex interactions between these two processes within the symbiotic organ. Here, we discuss recent results on the interplay between immunity and senescence in the nodule and the specificities of this relationship during legume-rhizobium symbiosis.
Sinorhizobium meliloti is a free-living, soil-dwelling bacterium that also engages in an N2-fixing symbiosis with leguminous plants of the genera Medicago, Melilotus, and Trigonella. It is aerobic and can be cultured in a variety of complex and defined minimal salts media, utilizing many sources of carbon, nitrogen, and phosphorus. Routes
for carbon metabolism include the Entner–Doudoroff and pentose phosphate pathways and the tricarboxylic acid cycle (TCA). S. meliloti and Medicago truncatula are important model organisms for rhizobia–legume symbiosis, whereby the bacteria occupy root nodules and receive nutrients from the plant in exchange for fixed N2 in the form of ammonia.
Abstract
The antimicrobial resistance crisis requires the introduction of novel antibiotics. The use of conventional broad-spectrum compounds selects for resistance in off-target pathogens and harms the microbiome. This is especially true for Mycobacterium tuberculosis, where treatment requires a 6-month course of antibiotics. Here we show that a novel antimicrobial from Photorhabdus noenieputensis, which we named evybactin, is a potent and selective antibiotic acting against M. tuberculosis. Evybactin targets DNA gyrase and binds to a site overlapping with synthetic thiophene poisons. Given the conserved nature of DNA gyrase, the observed selectivity against M. tuberculosis is puzzling. We found that evybactin is smuggled into the cell by a promiscuous transporter of hydrophilic compounds, BacA. Evybactin is the first, but likely not the only, antimicrobial compound found to employ this unusual mechanism of selectivity.
Abstract
BacA is a mycobacterial ATP-binding cassette (ABC) transporter involved in the translocation of water-soluble compounds across the lipid bilayer. Whole-cell-based assays have shown that BacA imports cobalamin as well as unrelated hydrophilic compounds such as the antibiotic bleomycin and the antimicrobial peptide Bac7 into the cytoplasm. Surprisingly, there are indications that BacA also mediates the export of different antibacterial compounds, which is difficult to reconcile with the notion that ABC transporters generally operate in a strictly unidirectional manner. Here we resolve this conundrum by developing a fluorescence-based transport assay to monitor the transport of cobalamin across liposomal membranes. We find that BacA transports cobalamin in both the import and export direction. This highly unusual bidirectionality suggests that BacA is mechanistically distinct from other ABC transporters and facilitates ATP-driven diffusion, a function that may be important for the evolvability of specific transporters, and may bring competitive advantages to microbial communities.
Bugs promote immune priming via gut-breaching bacteria
SIGNIFICANCE Immune priming has evolved in memory-deficient invertebrates to combat sudden infections effectively. Although this phenomenon has been reported in diverse insects, understanding of underpinning mechanisms remains limited. We found that a member of the gut microbiota in the bean bug Riptortus pedestris can cross the epithelia and be released into the hemolymph, thereby stimulating systemic immunity and preventing subsequent infection of lethal pathogens. In this process, both humoral and cellular immunity play a pivotal role, and there was no trade-off with insect fitness. This finding not only highlights the utilization of environmental microbes by insects for shaping immunity but also provides a unique, symbiotic perspective on bacterial intestinal barrier breaching, which has been generally discussed only in terms of pathology.
ABSTRACT Insects lack acquired immunity and were thought to have no immune memory, but recent studies reported a phenomenon called immune priming, wherein sublethal dose of pathogens or nonpathogenic microbes stimulates immunity and prevents subsequential pathogen infection. Although the evidence for insect immune priming is accumulating, the underlying mechanisms are still unclear. The bean bug Riptortus pedestris acquires its gut microbiota from ambient soil and spatially structures them into a multispecies and variable community in the anterior midgut and a specific, monospecies Caballeronia symbiont population in the posterior region. We demonstrate that a particular Burkholderia strain colonizing the anterior midgut stimulates systemic immunity by penetrating gut epithelia and migrating into the hemolymph. The activated immunity, consisting of a humoral and a cellular response, had no negative effect on the host fitness, but on the contrary protected the insect from subsequent infection by pathogenic bacteria. Interruption of contact between the Burkholderia strain and epithelia of the gut weakened the host immunity back to preinfection levels and made the insects more vulnerable to microbial infection, demonstrating that persistent acquisition of environmental bacteria is important to maintain an efficient immunity.
Abstract
The soil bacterium Sinorhizobium meliloti can establish a nitrogen fixing symbiosis with the model legume Medicago truncatula. The rhizobia induce the formation of a specialized root organ called nodule, where they differentiate into bacteroids and reduce atmospheric nitrogen into ammonia. Little is known on the mechanisms involved in nodule senescence onset and in bacteroid survival inside the infected plant cells. Whereas Toxin-Antitoxin (TA) systems have been shown to promote intracellular survival within host cells in human pathogenic bacteria, their role in symbiotic bacteria was rarely investigated. S. meliloti encodes several TA systems, mainly of the VapBC family. Here we present the functional characterization, through a multidisciplinary approach, of the VapBC10 TA system of S. meliloti. Following a MORE RNA-seq analysis, we demonstrated that the VapC10 toxin is an RNase that cleaves the anticodon loop of two tRNASer. Thereafter, a bioinformatics approach was used to predict VapC10 targets in bacteroids. This analysis suggests that toxin activation triggers a specific proteome reprogramming that could limit nitrogen fixation capability and viability of bacteroids. Accordingly, a vapC10 mutant induces a delayed senescence in nodules, associated to an enhanced bacteroid survival. VapBC10 TA system could contribute to S. meliloti adaptation to symbiotic lifestyle, in response to plant nitrogen status.
ABSTRACT
The ability to split one cell into two is fundamental to all life, and many bacteria can accomplish this feat several times per hour with high accuracy. Most bacteria call on an ancient homologue of tubulin, called FtsZ, to localize and organize the cell division machinery, the divisome, into a ring-like structure at the cell midpoint. The divisome includes numerous other proteins, often including an actin homologue (FtsA), that interact with each other at the cytoplasmic membrane. Once assembled, the protein complexes that comprise the dynamic divisome coordinate membrane constriction with synthesis of a division septum, but only after overcoming checkpoints mediated by specialized protein–protein interactions. In this Review, we summarize the most recent evidence showing how the divisome proteins of Escherichia coli assemble at the cell midpoint, interact with each other and regulate activation of septum synthesis. We also briefly discuss the potential of divisome proteins as novel antibiotic targets.
|
The 22nd International Conference on Nitrogen Fixation ICNF
And
The 20th Conference of the African Association for Biological Nitrogen Fixation AABNF
Abstract
Cobalamin (vitamin B12, herein referred to as B12) is an essential cofactor for most marine prokaryotes and eukaryotes1,2. Synthesized by a limited number of prokaryotes, its scarcity affects microbial interactions and community dynamics2,3,4. Here we show that two bacterial B12 auxotrophs can salvage different B12 building blocks and cooperate to synthesize B12. A Colwellia sp. synthesizes and releases the activated lower ligand α-ribazole, which is used by another B12 auxotroph, a Roseovarius sp., to produce the corrin ring and synthesize B12. Release of B12 by Roseovarius sp. happens only in co-culture with Colwellia sp. and only coincidently with the induction of a prophage encoded in Roseovarius sp. Subsequent growth of Colwellia sp. in these conditions may be due to the provision of B12 by lysed cells of Roseovarius sp. Further evidence is required to support a causative role for prophage induction in the release of B12. These complex microbial interactions of ligand cross-feeding and joint B12 biosynthesis seem to be widespread in marine pelagic ecosystems. In the western and northern tropical Atlantic Ocean, bacteria predicted to be capable of salvaging cobinamide and synthesizing only the activated lower ligand outnumber B12 producers. These findings add new players to our understanding of B12 supply to auxotrophic microorganisms in the ocean and possibly in other ecosystems.
Abstract
Specialized host–microbe symbioses canonically show greater diversity than expected from simple models, both at the population level and within individual hosts. To understand how this heterogeneity arises, we utilize the squash bug, Anasa tristis, and its bacterial symbionts in the genus Caballeronia. We modulate symbiont bottleneck size and inoculum composition during colonization to demonstrate the significance of ecological drift, the noisy fluctuations in community composition due to demographic stochasticity. Consistent with predictions from the neutral theory of biodiversity, we found that ecological drift alone can account for heterogeneity in symbiont community composition between hosts, even when 2 strains are nearly genetically identical. When acting on competing strains, ecological drift can maintain symbiont genetic diversity among different hosts by stochastically determining the dominant strain within each host. Finally, ecological drift mediates heterogeneity in isogenic symbiont populations even within a single host, along a consistent gradient running the anterior-posterior axis of the symbiotic organ. Our results demonstrate that symbiont population structure across scales does not necessarily require host-mediated selection, as it can emerge as a result of ecological drift acting on both isogenic and unrelated competitors. Our findings illuminate the processes that might affect symbiont transmission, coinfection, and population structure in nature, which can drive the evolution of host–microbe symbioses and microbe–microbe interactions within host-associated microbiomes.
Abstract
The α-Proteobacteria belonging to Bradyrhizobium genus are microorganisms of extreme slow growth. Despite their extended use as inoculants in soybean production, their physiology remains poorly characterized. In this work, we produced quantitative data on four different isolates: B. diazoefficens USDA110, B. diazoefficiens USDA122, B. japonicum E109 and B. japonicum USDA6 which are representative of specific genomic profiles. Notably, we found conserved physiological traits conserved in all the studied isolates: (i) the lag and initial exponential growth phases display cell aggregation; (ii) the increase in specific nutrient concentration such as yeast extract and gluconate hinders growth; (iii) cell size does not correlate with culture age; and (iv) cell cycle presents polar growth. Meanwhile, fitness, cell size and in vitro growth widely vary across isolates correlating to ribosomal RNA operon number. In summary, this study provides novel empirical data that enriches the comprehension of the Bradyrhizobium (slow) growth dynamics and cell cycle.
ABSTRACT Biomolecular condensates, such as the nucleoli or P-bodies, are non-membrane-bound assemblies of proteins and nucleic acids that facilitate specific cellular processes. Like eukaryotic P-bodies, the recently discovered bacterial ribonucleoprotein bodies (BR-bodies) organize the mRNA decay machinery, yet the similarities in molecular and cellular functions across species have been poorly explored. Here, we examine the functions of BR-bodies in the nitrogen-fixing endosymbiont Sinorhizobium meliloti, which colonizes the roots of compatible legume plants. Assembly of BR-bodies into visible foci in S. meliloti cells requires the C-terminal intrinsically disordered region (IDR) of RNase E, and foci fusion is readily observed in vivo, suggesting they are liquid-like condensates that form via mRNA sequestration. Using Rif-seq to measure mRNA lifetimes, we found a global slowdown in mRNA decay in a mutant deficient in BR-bodies, indicating that compartmentalization of the degradation machinery promotes efficient mRNA turnover. While BR-bodies are constitutively present during exponential growth, the abundance of BR-bodies increases upon cell stress, whereby they promote stress resistance. Finally, using Medicago truncatula as host, we show that BR-bodies enhance competitiveness during colonization and appear to be required for effective symbiosis, as mutants without BR-bodies failed to stimulate plant growth. These results suggest that BR-bodies provide a fitness advantage for bacteria during infection, perhaps by enabling better resistance against the host immune response.
SIGNIFICANCE While eukaryotes often organize their biochemical pathways in membrane-bound organelles, bacteria generally lack such subcellular structures. Instead, membraneless compartments called biomolecular condensates have recently been found in bacteria to enhance biochemical activities. Bacterial ribonucleoprotein bodies (BR-bodies), as one of the most widespread biomolecular condensates identified to date, assemble the mRNA decay machinery via the intrinsically disordered regions (IDRs) of proteins. However, the implications of such assemblies are unclear. Using a plant-associated symbiont, we show that the IDR of its mRNA degradation protein is necessary for condensate formation. Absence of BR-bodies results in slower mRNA decay and ineffective symbiosis, suggesting that BR-bodies play critical roles in regulating biochemical pathways and promoting fitness during host colonization.
Bradyrhizobium japonicum (including Bradyrhizobium diazoefficiens), a Gram-negative Alphaproteobacterium, is a member of the paraphyletic group of nitrogen-fixing bacteria
called rhizobia. Rhizobia live as free-living soil organisms or in symbiosis with leguminous plants. These symbioses are specific, with a rhizobial species able to associate with one or a limited number of plant hosts. The symbiosis between B. japonicum
and soybean (Glycine max) is important to agriculture because bacterial nitrogen fixation mitigates the need for chemical nitrogen fertilizer and allows the plant to grow in nitrogen-deficient soils.
HIGHLIGHTS
•Unipolar growth relies on a conserved set of genes distinct from the Rod system.
•PBP1a is the primary driver of unipolar peptidoglycan biosynthesis.
•Growth polarity is established by RgsS, an FtsN-like protein.
•Unipolar growth coordinates insertion of peptidoglycan, OMPs, and LPS at the growth pole.
ABSTRACT
The governing principles and suites of genes for lateral elongation or incorporation of new cell wall material along the length of a rod-shaped cell are well described. In contrast, relatively little is known about unipolar elongation or incorporation of peptidoglycan at one end of the rod. Recent work in three related model systems of unipolar growth (Agrobacterium tumefaciens, Brucella abortus, and Sinorhizobium meliloti) has clearly established that unipolar growth in the Hyphomicrobiales order relies on a set of genes distinct from the canonical elongasome. Polar incorporation of envelope components relies on homologous proteins shared by the Hyphomicrobiales, reviewed here. Ongoing and future work will reveal how unipolar growth is integrated into the alphaproteobacterial cell cycle and coordinated with other processes such as chromosome segregation and cell division.
Abstract
Mycobacterium tuberculosis (Mtb) infection is notoriously difficult to treat. Treatment efficacy is limited by Mtb’s intrinsic drug resistance, as well as its ability to evolve acquired resistance to all antituberculars in clinical use. A deeper understanding of the bacterial pathways that influence drug efficacy could facilitate the development of more effective therapies, identify new mechanisms of acquired resistance, and reveal overlooked therapeutic opportunities. Here we developed a CRISPR interference chemical-genetics platform to titrate the expression of Mtb genes and quantify bacterial fitness in the presence of different drugs. We discovered diverse mechanisms of intrinsic drug resistance, unveiling hundreds of potential targets for synergistic drug combinations. Combining chemical genetics with comparative genomics of Mtb clinical isolates, we further identified several previously unknown mechanisms of acquired drug resistance, one of which is associated with a multidrug-resistant tuberculosis outbreak in South America. Lastly, we found that the intrinsic resistance factor whiB7 was inactivated in an entire Mtb sublineage endemic to Southeast Asia, presenting an opportunity to potentially repurpose the macrolide antibiotic clarithromycin to treat tuberculosis. This chemical-genetic map provides a rich resource to understand drug efficacy in Mtb and guide future tuberculosis drug development and treatment.
Highlights
•Trachea expose phosphatidylserine, making them sensitive to AMPs
•Turandots are secreted proteins that protect the respiratory epithelium
•Turandots shield phosphatidylserine, preventing pore formation by AMPs
•Turandots favor Drosophila resilience to stress by preventing immunopathology
Summary
7An efficient immune system must provide protection against a broad range of pathogens without causing excessive collateral tissue damage. While immune effectors have been well characterized, we know less about the resilience mechanisms protecting the host from its own immune response. Antimicrobial peptides (AMPs) are small, cationic peptides that contribute to innate defenses by targeting negatively charged membranes of microbes. While protective against pathogens, AMPs can be cytotoxic to host cells. Here, we reveal that a family of stress-induced proteins, the Turandots, protect the Drosophila respiratory system from AMPs, increasing resilience to stress. Flies lacking Turandot genes are susceptible to environmental stresses due to AMP-induced tracheal apoptosis. Turandot proteins bind to host cell membranes and mask negatively charged phospholipids, protecting them from cationic pore-forming AMPs. Collectively, these data demonstrate that Turandot stress proteins mitigate AMP cytotoxicity to host tissues and therefore improve their efficacy.
ABSTRACT
Small, antimicrobial peptides are often produced by eukaryotes to control bacterial populations in both pathogenic and mutualistic symbioses. These include proline-rich mammalian immune peptides and cysteine-rich peptides produced by legume plants in symbiosis with rhizobia. The fitness of the bacterial partner is dependent upon their ability to persist in the presence of these antimicrobial peptides. In the case of Escherichia coli and Mycobacterium tuberculosis pathogens and nitrogen-fixing legume symbionts (rhizobia), the ability to survive exposure to these peptides depends on peptide transporters called SbmA (also known as BacA) or BclA (for BacA-like). However, how broadly these transporters are distributed amongst bacteria, and their evolutionary history, is poorly understood. Here, we used hidden Markov models, phylogenetic analysis, and sequence similarity networks to examine the distribution of SbmA/BacA and BclA proteins across a representative set of 1,255 species from across the domain Bacteria. We identified a total of 71 and 177 SbmA/BacA and BclA proteins, respectively. Phylogenetic and sequence similarity analyses suggest that these protein families likely did not evolve from a common ancestor and that their functional similarity is instead a result of convergent evolution. In vitro sensitivity assays using the legume peptide NCR247 and several of the newly-identified BclA proteins confirmed that transport of antimicrobial peptides is a common feature of this protein family. Analysis of the taxonomic distribution of these proteins showed that SbmA/BacA orthologs were encoded only by species in the phylum Pseudomonadota and that they were primarily identified in just two orders: Hyphomicrobiales (class Alphaproteobacteria) and Enterobacterales (class Gammaproteobacteria). BclA orthologs were somewhat more broadly distributed and were found in clusters across four phyla. These included several orders of the phyla Pseudomonadota and Cyanobacteriota, as well as the order Mycobacteriales (phylum Actinomycetota) and the class Negativicutes (phylum Bacillota). Notably, many of the clades enriched for species encoding BacA or BclA orthologs also include many species known to interact with eukaryotic hosts in mutualistic or pathogenic interactions. Collectively, these observations suggest that SbmA/BacA and BclA proteins have been repeatedly co-opted to facilitate both mutualistic and pathogenic associations with eukaryotic hosts by allowing bacteria to cope with host-encoded antimicrobial peptides.
Significance
Symbiosis between legumes and nitrogen-fixing bacteria is dependent on the Symbiosis receptor-like kinase (SYMRK). Phosphorylation is an important signal for pathway activation, and in this study, we mapped essential phosphorylation sites in the intracellular part of SYMRK. We identify a conserved “alpha-I” motif in SYMRK and demonstrate that phosphorylation of four serine residues in the alpha-I motif drives the cellular program leading to root nodule formation.
Abstract
Symbiosis receptor-like kinase SYMRK is required for root nodule symbiosis between legume plants and nitrogen-fixing bacteria. To understand symbiotic signaling from SYMRK, we determined the crystal structure to 1.95 Å and mapped the phosphorylation sites onto the intracellular domain. We identified four serine residues in a conserved “alpha-I” motif, located on the border between the kinase core domain and the flexible C-terminal tail, that, when phosphorylated, drives organogenesis. Substituting the four serines with alanines abolished symbiotic signaling, while substituting them with phosphorylation-mimicking aspartates induced the formation of spontaneous nodules in the absence of bacteria. These findings show that the signaling pathway controlling root nodule organogenesis is mediated by SYMRK phosphorylation, which may help when engineering this trait into non-legume plants.
HIGHLIGHTS
•Large-scale phenotyping of E. coli cell morphology, cell cycle events, and growth
•Extensive phenotypic plasticity of gene deletions across nutrient conditions
•Emergence of 5 robust cell cycle laws across genetic and metabolic perturbations
•Population-level integration of cell cycle processes and growth rate with cell size
SUMMARY
To examine how bacteria achieve robust cell proliferation across diverse conditions, we developed a method that quantifies 77 cell morphological, cell cycle, and growth phenotypes of a fluorescently labeled Escherichia coli strain and >800 gene deletion derivatives under multiple nutrient conditions. This approach revealed extensive phenotypic plasticity and deviating mutant phenotypes were often nutrient dependent. From this broad phenotypic landscape emerged simple and robust unifying rules (laws) that connect DNA replication initiation, nucleoid segregation, FtsZ ring formation, and cell constriction to specific aspects of cell size (volume, length, or added length) at the population level. Furthermore, completion of cell division followed the initiation of cell constriction after a constant time delay across strains and nutrient conditions, identifying cell constriction as a key control point for cell size determination. Our work provides a population-level description of the governing principles by which E. coli integrates cell cycle processes and growth rate with cell size to achieve its robust proliferative capability.
|
 Your new post is loading...
Your new post is loading...