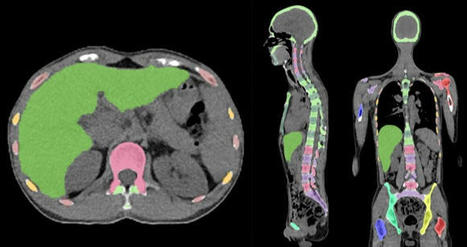
Researchers investigating the benefits of 3D printing technology found it can deliver significant improvements to the running of hospitals.
The research, which compared the drawbacks and advantages of using 3D printing technology in hospitals, has been published in the International Journal of Operations and Production Management.
The study revealed that introducing such technology into hospitals could help alleviate many of the strains the UK healthcare system and healthcare systems worldwide face.
Boosting surgery success rates
- 3D printing makes it possible for surgical teams to print 3D models based on an individual patient’s surgical needs, providing more detailed and exact information for the surgeon to plan and practice the surgery, minimising the risk of error or unexpected complications.
- the use of 3D printed anatomical models was useful when communicating the details of the surgery with the patient, helping to increase their confidence in the procedure.
Speeding up patient recovery time
- significant reduction in post-surgery complications, patient recovery times and the need for subsequent hospital appointments or treatments.
Speeding up procedures
- provide surgeons with custom-built tools for each procedure, with the findings revealing that surgeries with durations of four to eight hours were reduced by 1.5 to 2.5 hours when patient-specific instruments were used.
- could also make surgeries less invasive (for example, removing less bone or tissue)
- result in less associated risks for the patient (for example, by requiring less anaesthesia).
Real-life training opportunities
- enables trainee surgeons to familiarise themselves with the steps to take in complex surgeries by practicing their skills on examples that accurately replicate real patient problems, and with greater variety.
Careful consideration required
Despite the research showing strong and clear benefits of using 3D printing, Dr Chaudhuri and his fellow researchers urge careful consideration for the financial costs.
3D printing is a significant financial investment for hospitals to make. In order to determine whether such an investment is worthwhile, the researchers have also developed a framework to aid hospital decision-makers in determining the return on investment for their particular institution.
read more at https://www.healtheuropa.eu/3d-printing-technology-boosts-hospital-efficiency-and-eases-pressures/108544/
Lire l'article complet sur : www.healtheuropa.eu
Via nrip



 Your new post is loading...
Your new post is loading...